AAA Quantitation and Data Analysis
Amino Acid Analysis Assays
We offer a range of assays for amino acid analysis, tailored to your sample type and specific requirements. Here’s a breakdown of our processes and the amino acids we report:
Purified Proteins (Acid Vapor-Phase Hydrolysis)
For purified proteins undergoing acid vapor-phase hydrolysis (6 N HCl), we report the analysis of 16 naturally occurring amino acids.
- Asparagine (Asn) and Glutamine (Gln) are deamidated to their respective acids during hydrolysis and are reported as ASX (Asparagine/Asparagic Acid) and GLX (Glutamine/Glutamic Acid).
- Cysteine is destroyed during hydrolysis but can be preserved if alkylated before the process. Please ensure you request Cysteine analysis on your sample submittal form if it is essential. Note that this requires a separate assay and incurs an additional charge due to the use of a different detector.
- Tryptophan is destroyed during HCl hydrolysis, so it requires a separate assay using Methanesulfonic Acid hydrolysis. This assay demands a larger sample size and may have some variability.
- Isoleucine (ILE) can sometimes be challenging to hydrolyze, and it may report lower than expected, especially if it is adjacent to other branched amino acids in the sequence (such as Isoleucine or Valine). This is a known issue that can affect the results.
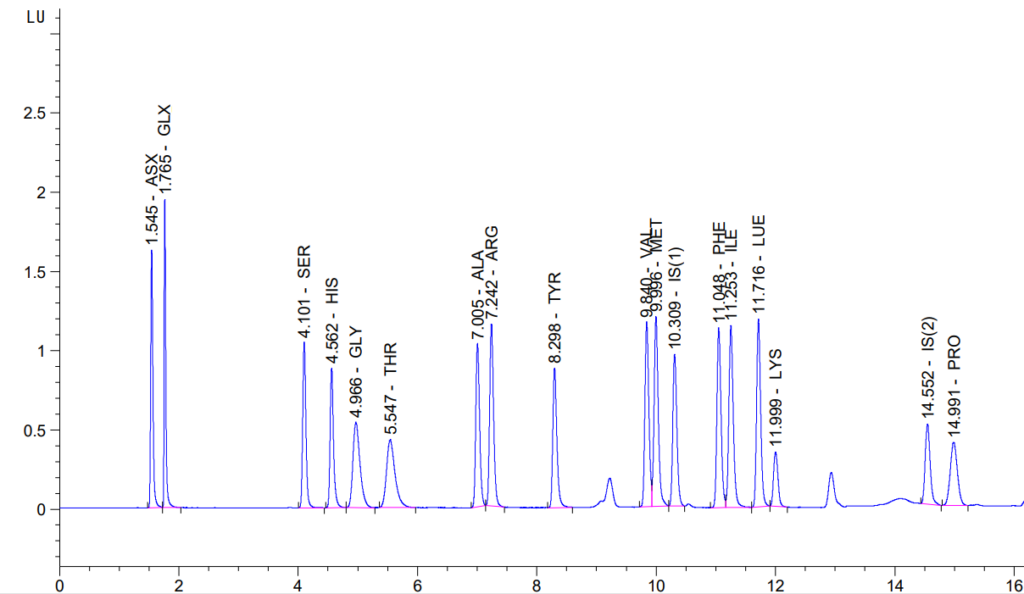
Solid Samples (e.g., Feed or Meal)
Solid samples are processed via acid liquid-phase hydrolysis (6 N HCl), and we will report the 16 naturally occurring amino acids. Please note that Cysteine and Tryptophan are not preserved in this process and will not be reported.
Physiological Samples (e.g., Serum)
For physiological samples like serum, we report the 19 naturally occurring amino acids. In addition, we can include the following amino acids upon request:
- Citrulline (Cit)
- Beta-Alanine (ß-Ala)
- Taurine (Tau)
- Ornithine (Orn)
Please specify these additional amino acids on your request form if needed.
The Excel Workbook – Results Summary
After analysis, you will receive a detailed Excel Workbook summarizing the results, which includes:
- Replicates Averaged: The data from multiple runs is averaged for accuracy.
- Percent Composition: Calculated for each amino acid in your sample.
- Sample Amount: The amount of sample in the aliquot is calculated in several ways, allowing for flexibility in how you view your results.
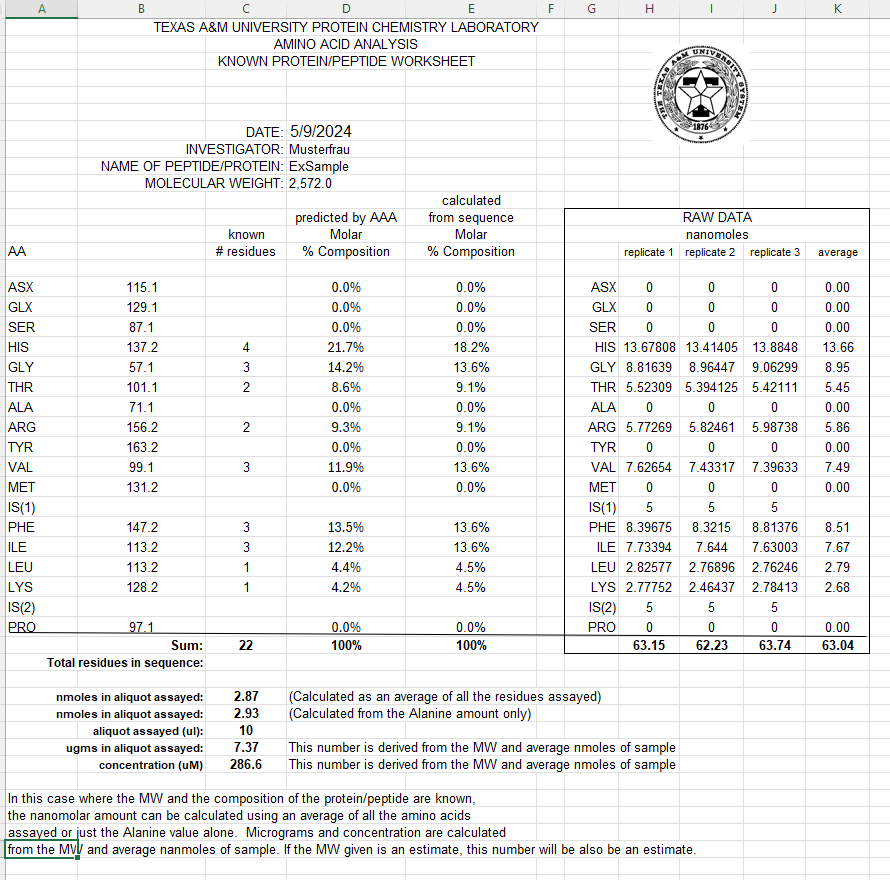
If you provide the molecular weight and sequence of the protein, we can calculate the amount of protein in nanomoles and compare the known molar percent composition to the results obtained from the assay.
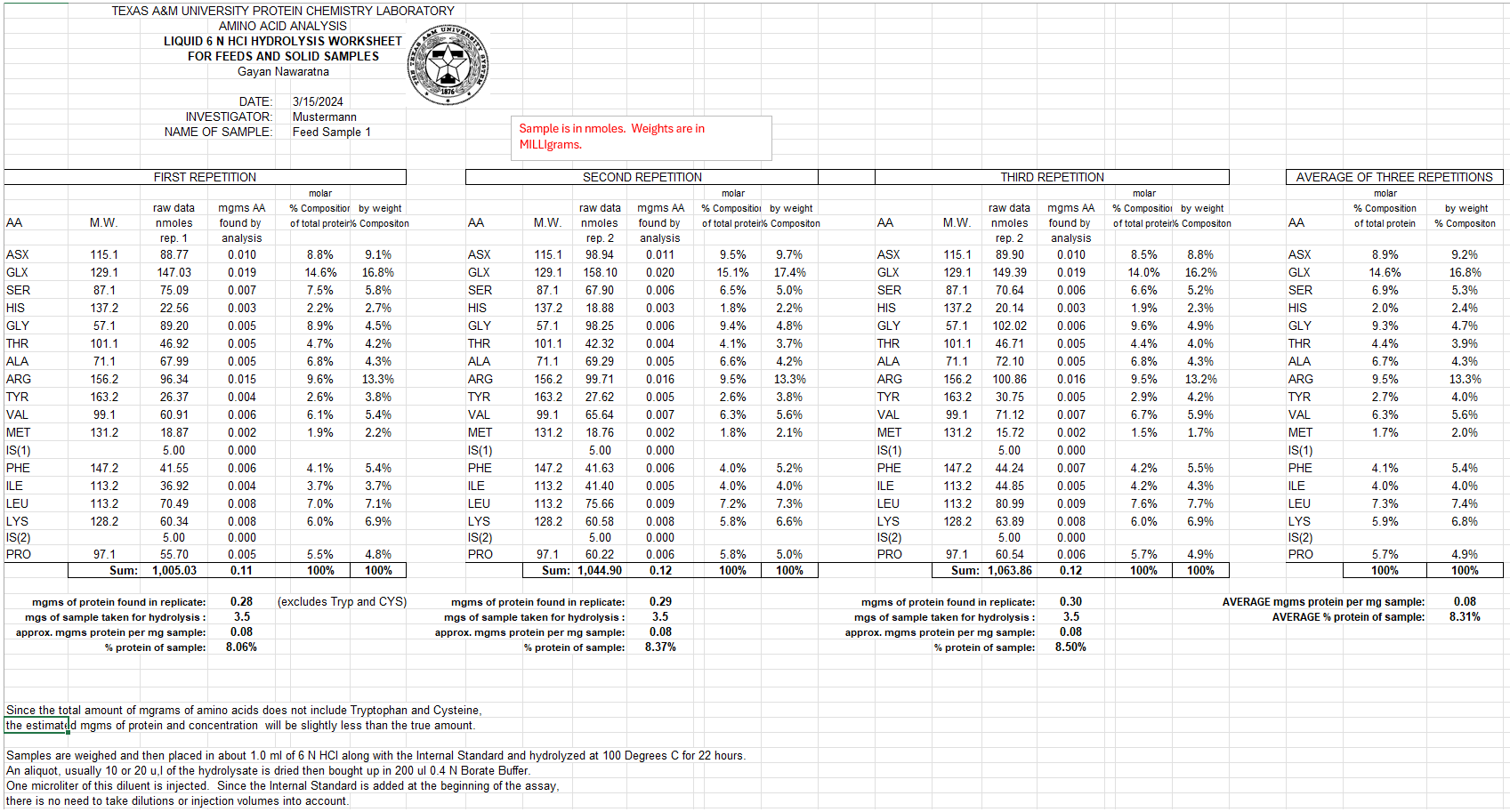
For quantitative accuracy, we recommend running your samples in triplicate. Please note there is an additional charge for triplicates or duplicates, and it’s up to the investigator to specify the preferred number of replicates.
Data Retention
- The raw data is kept electronically for 1 year.
- The summary data in the Workbook is retained for 3 years.
This ensures that your data is securely stored and easily accessible for future reference or analysis.
For any specific requests or to clarify the details of your sample analysis, please don’t hesitate to contact us!
